On the evening of May 23, Professor GAO Shaorong/GAO Yawei’s research team from the School of Life Sciences and Technology of Tongji University and Professor SHEN Bin’s team from Nanjing Medical University jointly published online in nature cell biology a paper entitled "N6-methyladenosine regulates maternal RNA maintenance in oocytes and timely RNA decay during mouse maternal-to-zygotic transition". The dynamic changes of m6A modification on RNA (gene transcripts) as the first key event in mouse early embryonic development - maternal zygotic transition was discovered for the first time. It also revealed the unique molecular mechanism of m6A regulating the stability of maternal gene transcripts and the timely degradation of 2-cell-specific gene transcripts. The team draw the first dynamic map of RNA m6A modification during early mouse embryonic development with the help of the improved micro-cell initiated sequencing technology. This was the third time that the team's research results were published in leading international academic journals within one month.

The paper published on nature cell biology
The initial stage of early embryonic development is the formation of fertilized eggs by oocytes through fertilization. Fertilization triggers the degradation of ribonucleic acids and proteins stored in oocytes, and the zygotic genome activation (ZGA). This process is also known as the maternal to zygotic transition (MZT) process. There are many levels of reprogramming in the process of maternal zygote transition, such as proteome, histone apparent modification group, DNA methylation group and transcriptome. With the development of single-cell sequencing technology, important achievements have been made in transcriptome in recent years. However, due to the limitations of technology development, there is still much to be done in the research on RNA modification distribution and regulation mechanism.
Genetic information is stored in DNA in organisms, and RNA, i.e. transcripts, is generated through transcription, among which some transcripts will be directly translated into proteins to function (coding RNA/ transcripts), and other transcripts participate in the regulation function in cells in the form of RNA (non-coding RNA/ transcripts). The chemical modification on RNA is considered to be the basis of regulation at many RNA levels. At present, more than 200 kinds of RNA chemical modifications have been found, among which N6 methyladenine (m6A) is the most abundant modification on eukaryotic mRNA, and is widely considered to be involved in the regulation of RNA splicing, transport, degradation, translation and phase separation. The establishment of m6A modification on RNA depends on methyltransferase protein complexes, including METTL3, METTL14, METTL16, WTAP and KIAA1429. Studies have found that lack of these proteins can lead to the abnormal differentiation of mouse embryonic stem cells, oogenesis and early embryonic development. However, the traditional m6A modified sequencing technology requires a large amount of RNA as raw materials, while the early mouse embryo materials are very scarce. Therefore, there has been a shortage in the dynamic detection of m6A modified histomics in the early embryonic development of mice and other mammals, which limits the development of related mechanism research.
Researchers started from establishing a ULI-MeRIP SEQ suitable for 50NG total RNA initiation by optimizing the traditional m6A sequencing to reveal the omics dynamics of m6A in early mouse embryos. This approach can accurately detect the distribution position of m6A modifier on RNA and highly overlap with the m6A modification information obtained from a large number of RNA initiation. The research team found out m6A modification of oocytes and embryonic materials involved in the process of mother zygote transformation, such as GV oocytes (late meiotic biphasic stage), MII oocytes (middle stage of second meiosis, mature oocytes), fertilized eggs and early cleavage embryos (2-cell and 4-cell embryos). The results showed that about 40% of maternal transcripts were enriched with m6A modification. The expression of these transcripts in oocytes was generally higher than that of unmodified RNA, and most of them were degraded after fertilization. About 70% of the ZGA transcripts activated after fertilization were enriched with m6A modification, including 2-cell-specific RNA such as Zscan4 (Fig. 1). It is worth noting that many specific noncoding transcripts at the developmental stage, such as the high abundance of MTA in oocytes (belonging to the MaLR family) and the 2-cell-specific expression of MERVL (belonging to the ERVL family), are highly enriched in m6A modification.
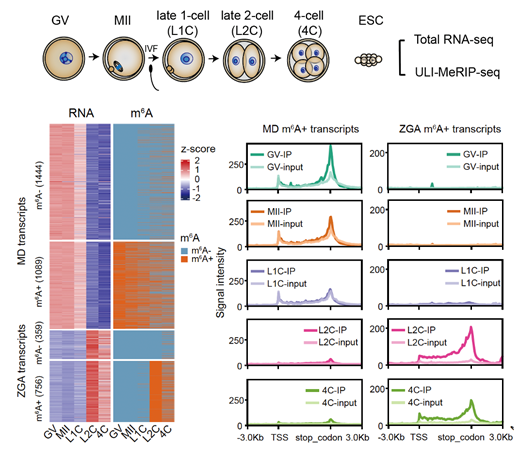
Figure 1. Distribution and enrichment of m6A during parent zygote transition
In their attempt to reveal the regulatory effect of m6A on maternal and progeny transcripts, the researchers used the mouse model of oocyte specific knockout of Kiaa1429 (one of the members of RNA methyltransferase complex) and the methyltransferase METTL3 inhibitor STM2457 to obtain the eggs and embryos modified by m6A to establish the defect, respectively. The research team discovered that the maternal transcripts, including the transcripts of the maternal transposon MTA, showed a significant decrease in their own abundance after lack of m6A modification (Fig. 2); Interestingly, inhibiting the establishment of m6A on ZGA transcripts would trigger transcripts that were transiently expressed in the 2-4-cell phase, such as Zscan4, and the retrotransposon MERVL could not be degraded in time, thus causing the arrest of egg and embryo development respectively. These results showed that m6A modification on transcripts regulated the maintenance of parental transcripts and the orderly degradation of progeny transcripts, ensuring the smooth occurrence of maternal progeny transition in early life.
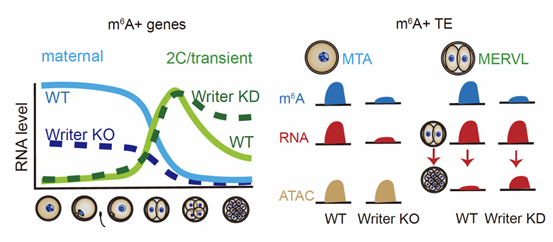
Figure 2. M6A regulation of maternal zygote transition in mice
According to GAO Shaorong, the team established the dynamic map of m6A modification on transcripts during early embryonic development of mice for the first time by high-throughput sequencing, and confirmed that the maternal transposon MTA and 2-cell transposon MERVL were highly enriched in m6A modification. It was further found that m6A modification maintained the stability of maternal RNA in oocytes and the timely degradation of transcripts transiently expressed after fertilization. The different regulatory functions of m6A modification in oocyte and zygote gene activation reflected the complexity of regulation in the initial stage of life, and open up a new window for further exploring the mystery of life regulation.
The first authors of this paper are WU You and XU Xiaocui, postdoctoral research fellows of Tongji University, and PhD student QI Meijie of Nanjing Medical University while GAO Shaorong, GAO Yawei and SHEN Bin the corresponding authors. Dr. CHEN Chuan from the Obstetrics and Gynecology Hospital of Zhejiang University, Professor YI Chengqi from Peking University, Dr. XIANG Junhong from the University of Chicago, and Professor LIU Hongbin from the Reproductive Medicine Research Center of Shandong University participated in this research. The research was supported by the Ministry of Science and Technology, the National Natural Science Foundation of China and the Shanghai Science and Technology Commission.
Link to paper: https://www.nature.com/articles/s41556-022-00915-x
The research team has been committed to research on analyzing the m6A regulation in early mouse embryos and post implantation embryonic development and embryonic stem cells for decades. Their research on FTO gene regulation of nuclear appearance and development by regulating m6A modification of Line1 RNA was published online in Science on May 5, 2022. The YTHDC1 regulation of the function of LINE1 RNA in early embryos and embryonic stem cells was also proposed by the team.
GAO Shaorong's team is committed to analysis of the apparent regulation in early mouse embryonic development and cell reprogramming. Their research results, in cooperation with ZHANG Yong of the School of Life Science and Technology, on the development process of mouse preimplantation embryos were introduced in a paper entitled "the region rich in binuclear glycine (CPG) jointly marked by allele specific H3K9me3 and DNA methylation as the potential imprinting control region in preimplantation embryos" in nature cell biology on April 28, 2022. Another paper entitled “FTO mediates LINE1 m6A demethylation and chromatin regulation in mSECs and mouse development” was published online, jointly with HE Chuan’s team of the University of Chicago, in science on May 5 this year, demonstrating the molecular mechanism of FTO gene regulating nuclear appearance and development by regulating m6A modification of LINE1 RNA.
Sources:
https://news.tongji.edu.cn/info/1003/80906.htm
https://news.tongji.edu.cn/info/1003/80828.htm