On September 14, 2020, Gao’ lab in the Life Science and Technology College of Tongji University have published “Precise allele-specific genome editing by spatiotemporal control of CRISPR-Cas9 via pronuclear transplantation”. In this study, researchers established a new gene-editing method “Past-CRISPR (parental allele-specific gene-targeting)” that could edit parental allele sites specifically and reduce mosaicism significantly in gene-editing animal models.
Recently, CRISPR-Cas9 has developed into the system that can efficiently edit gene sequences within the genome. The gene-editing model animals can be efficiently generated through the co-injection of Cas9 and sgRNAs into the zygote or oocytes. Nonetheless, the constitutively active nature of Cas9 makes it difficult to achieve precise spatial and temporal control over Cas9 gene-editing activity, which leads to numerous limitations and unpredictable hazards. First, the majority of generated gene-edited embryos showed mosaic genotypes due to prolonged Cas9 on-targeting activity after zygotic division in the traditional injection methods, which needs to be resolved by long-time reproductive processes. Second, monoallelic site editing is ineffective under the traditional CRISPR-Cas9 system because the cleavage efficiency of the CRISPR-Cas9 system is spatiotemporally uncontrollable.
To spatiotemporally control Cas9 activity and achieve the precise allele-specific gene editing, researchers proposed two assumes: First, the zygotic pronucleus stage is the unique window where the parental genomes are physically separated across the life cycle, probably giving us a space to achieve parental allele-specific genome targeting. Second, the amounts of Cas9 and sgRNAs can be diluted through degradation following pronuclear transplantation into zygotes, which might achieve spatiotemporally controlled Cas9 activity.
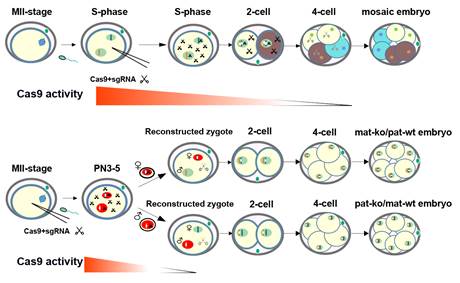
Based on the two assumes, researchers co-injected Cas9 mRNA and sgRNAs into MII stage oocytes and then performing in vitro fertilization (IVF). When zygotes developed into PN 3-4 stage (7-9 h.p.f), they isolated paternal or maternal pronucleus of the zygote from the injection group and transferred them into wild-type embryos that had been depleted of paternal or maternal pronucleus, respectively, which generated reconstructed normal diploid zygotes. Theoretically, in the reconstructed zygote, one of the two sets of DNA had been edited, and the other was wild type (Pat-ko/Mat-wt or Mat-ko/Pat-wt).
To test the efficiency of Past-CRISPR methods in multiple gene sites, researchers demonstrated that Past-CRISPR methods can remain the normal editing efficiency (85.7%) and have high monoallelic targeting efficiency in each gene site (91.7%). More importantly, parental allele-specific sites can be selectively recognized and modified by Cas9 with the help of Past-CRISPR methods. Meanwhile, these parental allele-specific edited embryos exhibited significantly reduced mosaicism compared with that of the traditional injection methods (7.1% vs 84.6%).
Then, researchers used Past-CRISPR methods to widely improve various genome editing applications. The methods can be particularly useful for verification of function and phenotype of imprinting genes, and rapidly generating viable monoallelic targeting animal models with embryonically lethal mutations, which can be transmitted to F1 generations by crossing. Meanwhile,the methods can generate complete gene knockout animal models without genotypic mosaicism in a one-step process. Lastly, Past-CRISPR can be used for treating dominant genetic diseases. Researchers focused on a mouse model with a dominant point mutation in Fgfr3 (Fgfr3 Gly369Cys mutation), causing dwarfism with features mimicking human achondroplasia. And they found the dominant allelic loci could be selectively disrupted by Past-CRISPR methods, then the aberrant bone phenotype can be blocked and corrected in the next generation.
source:https://news.tongji.edu.cn/info/1002/74957.htm
https://www.nature.com/articles/s41467-020-18391-y.pdf